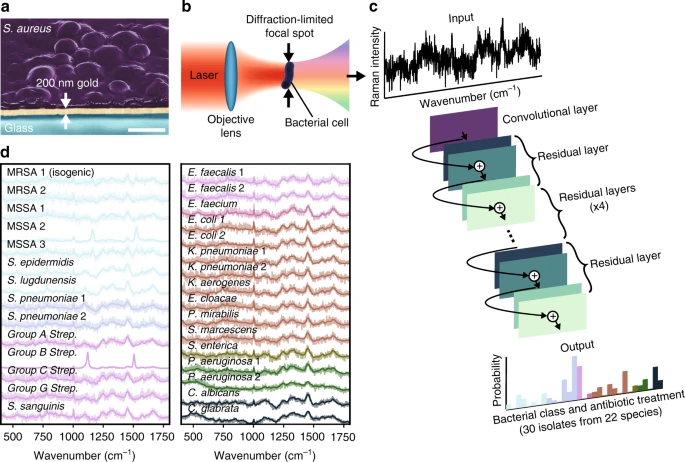
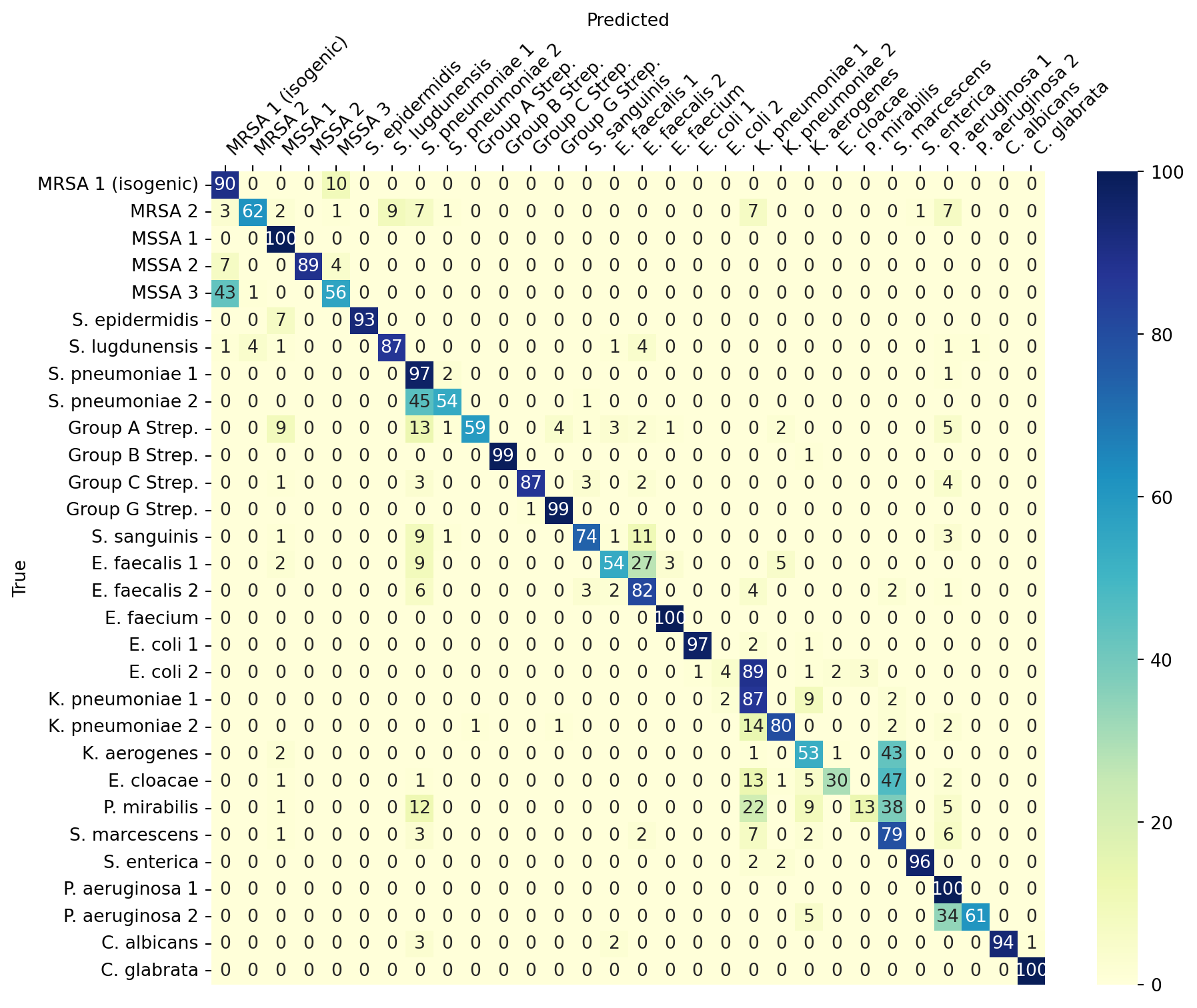
ResNet(
(conv1): Conv1d(1, 64, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn1): BatchNorm1d(64, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(encoder): Sequential(
(0): Sequential(
(0): ResidualBlock(
(conv1): Conv1d(64, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential(
(0): Conv1d(64, 100, kernel_size=(1,), stride=(1,), bias=False)
(1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential()
)
)
(1): Sequential(
(0): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(2,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential(
(0): Conv1d(100, 100, kernel_size=(1,), stride=(2,), bias=False)
(1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential()
)
)
(2): Sequential(
(0): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(2,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential(
(0): Conv1d(100, 100, kernel_size=(1,), stride=(2,), bias=False)
(1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential()
)
)
(3): Sequential(
(0): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(2,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential(
(0): Conv1d(100, 100, kernel_size=(1,), stride=(2,), bias=False)
(1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential()
)
)
(4): Sequential(
(0): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(2,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential(
(0): Conv1d(100, 100, kernel_size=(1,), stride=(2,), bias=False)
(1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential()
)
)
(5): Sequential(
(0): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(2,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential(
(0): Conv1d(100, 100, kernel_size=(1,), stride=(2,), bias=False)
(1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
)
)
(1): ResidualBlock(
(conv1): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn1): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(conv2): Conv1d(100, 100, kernel_size=(5,), stride=(1,), padding=(2,), bias=False)
(bn2): BatchNorm1d(100, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(shortcut): Sequential()
)
)
)
(linear): Linear(in_features=3200, out_features=30, bias=True)
)